Scale up your NGS workflow for SARS-CoV-2 research and variant detection●
Efficient viral surveillance with Tecan's sensitive and rapid SARS-CoV-2 genome sequencing.
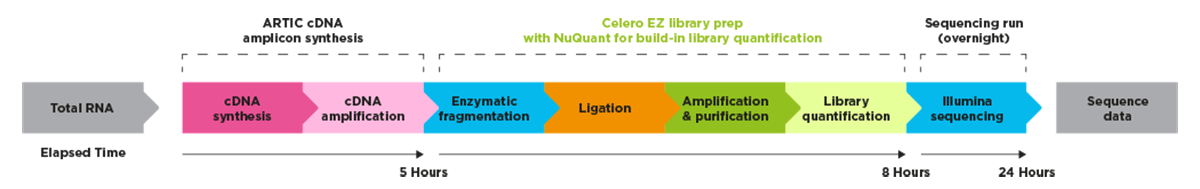
Genomic labs can be empowered with optimized automated protocols for library preparation.

More than just a platform, DreamPrep NGS offers a full
NGS solution for SARS-CoV-2 sequencing
- Single contact point for support and troubleshooting.
- Optimal performance with Tecan NGS reagents for faster and more efficient workflows.
- Intuitive and User-friendly interface empowers your daily operations.
- No sample loss in QC and normalization, which takes only 6 minutes.
- Longer walk-away times enabled by the on-deck thermocycler.
Testimonials Section
With this approach, we could already identify the presence of new variants in Neuchâtel in April 2020.
Dr. Nicolas Sierro, PMI science
How to identify the presence of SARS-CoV-2 virus variants in a certain area?
Watch the webinar with Dr. Nicolas Sierro, Manager Genomics & Transcriptomics at PMI Science, who will shed the light on this question using Neuchâtel in Switzerland as an example. This webinar highlights the process for identifying the presence of the VOC 20I/501Y.V1 variant in samples from the Neuchâtel area – from early RT-qPCR S-dropout screening to full viral genome sequencing.
- Peinetti AS, et al. (2021). Direct detection of human adenovirus or SARS-CoV-2 with ability to inform infectivity using DNA aptamer-nanopore sensors. Sci Adv. doi: 10.1126/sciadv.abh2848. Summary: Researchers here develop a “no sample pre-treatment” method for direct detection and differentiation of infectious from noninfectious human adenovirus and SARS-CoV-2 using DNA aptamers. Celero DNA-Seq Kit was used as the kit for library preparation.
- Wilkinson, et al. (2021). Genomic insights into early SARS-CoV-2 strains isolated in Reunion Island. medRxiv. doi: https://doi.org/10.1101/2021.01.21.21249623. [Preprint] Sumary: from the University of La Réunion performed in-house genome sequencing to understand the diffusion of SARS-Cov-2 strains in Reunion Island. The researchers used a hybrid approach, by combining Oxford Nanopore Technologies and Illumina sequencing, to improve the viral genomic assemblies. The libraries for Illumina sequencing were generated using 10 ng of cDNA using the Celero EZ library prep protocol.
