Keywords:
Drug discovery is a lengthy and expensive process, particularly in the early candidate identification stages, where screening of large numbers of compounds is required. Researchers at the University of Central Florida are using artificial intelligence (AI) to aid candidate selection for antimalarial drugs, making this selection process more efficient and cost effective, as well as increasing the likelihood of success.
Malaria is one of the biggest challenges facing the world today. Not only does it affect a considerable proportion of the world’s population, but the prevalence of drug-resistant parasites is causing the limited number of treatments available to become less effective. Researchers in the Burnett School of Biomedical Sciences at the University of Central Florida, Orlando, have turned to AI to assist in the discovery of new candidate drugs, initially focusing on malaria. Arash Keshavarzi Arshadi, a computational biotechnology PhD student, explained: “AI offers considerable benefits for drug discovery
in terms of time and cost savings, allowing researchers to be far more selective when choosing compound libraries to study. For example, the time and cost involved in physically screening a library of 250,000 compounds is simply prohibitive. However, using AI, we can screen all 250,000 compounds in silico to identify potential antimalarials – such as macrocyclic scaffolds – while excluding compounds that would definitely not be effective. This approach saves us time and money, as we only need to buy and screen the ‘hits’ identified by AI.”
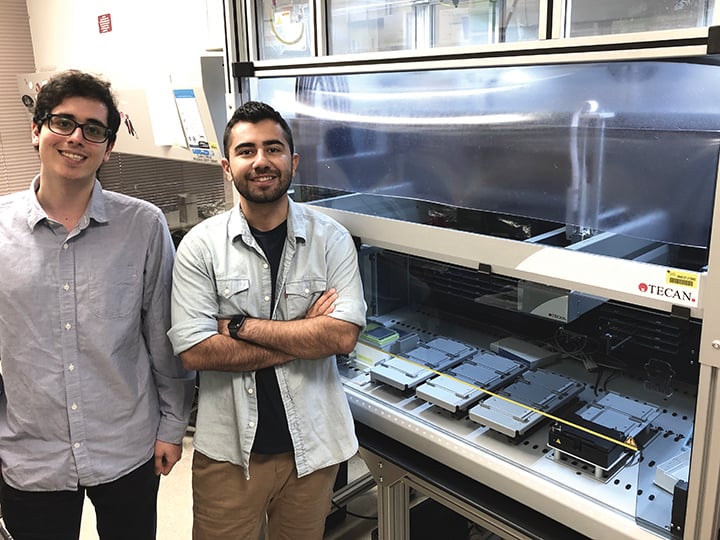 Milad Salem (left) and Arash Keshavarzi Arshadi
Milad Salem (left) and Arash Keshavarzi Arshadi
Milad Salem, an AI scientist in the Department of Computer Engineering, took up the story: “This work involves using a deep learning approach, finding and adapting computer models to predict the potential efficacy of drug candidates. We initially chose to study malaria, looking for novel candidates to overcome the growing issue of drug resistance. The idea was to establish a model that could successfully predict antimalarial candidates, before using this model to develop further algorithms for other diseases. From a computational standpoint, having the correct data to make the model is essential. Finding that data can be really challenging – sometimes impossible – but in this case we were lucky enough to have publicly available data from a major pharmaceutical company that had identified around 13,000 compounds active for a specific malarial strain. We used this dataset to train our algorithm to find hidden patterns in those compounds.”
Once the in silico model – known as DeepMalaria1 – had been trained, the next step was to test the accuracy of the algorithm using macrocyclic compound libraries. With large numbers of compounds to screen, turning to automation was an obvious decision. Arash continued: “We bought a number of compound libraries to test the algorithm, comparing the theoretical results predicted by AI with those observed during practical high throughput screening on a Freedom EVO® workstation in Dr Debopam Chakrabarti’s laboratory. This involved single-point screening of one micromolar solutions of 2,500 compounds, and confirmed that our algorithm predicted hits with an overall accuracy of about 87.5 percent for those that inhibited growth by 50 percent. This rose to 100 percent for nanomolar active compounds. In other words, DeepMalaria did not miss any highly potent macrocyclic antimalarials.”
“We then went on to determine the half maximal inhibitory concentration (IC50) of the hits we had identified, using the Freedom EVO to perform serial dilutions and transfer them to a black plate for SYBR® Green screening to assess cell viability of malaria parasites. Automating this high throughput screening is a very efficient way to perform our workflow; it would take so much time to do manually. I had no previous experience with this type of system, but found it straightforward to work with. I quickly learnt the basics, and it is easy to write scripts for our screening and toxicity assays in Freedom EVOware®. I also attended a Tecan workshop, where I picked up some useful tips, particularly for working with 384-well plates. Previously, we did everything in 96-well plates, but switching to a 384-well format has made our workflow more efficient and economical, reducing costs by over 50 percent.”
Milad added: “Normally, data that has already been physically screened is given to an AI model to see how good it is at predicting what has already happened. However, we wanted to save time by working prospectively, using our model to predict which compounds would be active against malaria, and we were really pleased with how well our AI performed. These results have given us hope that deep learning models can find patterns that humans can’t, and may be able to work in a more abstract way than was previously thought possible.”
“Moving forward, we plan to continue this work by developing more algorithms targeting other diseases, such as cancer and HIV, which Dr Jiann Shiun Yuan’s laboratory is studying. AI has huge potential to aid drug discovery, and having publicly available data for malaria really helped us to drive our research forward. We would encourage all researchers in this sector to take an open source approach to data sharing, allowing more diseases to benefit from AI,” Arash concluded.
1) Arshadi, A; Salem, M; et al. DeepMalaria: Artificial Intelligence Driven Discovery of Potent Antiplasmodials. Frontiers in Pharmacol, January 2020, DOI: 10.3389/fphar.2019.01526
Automating this high throughput screening is a very efficient way to perform our workflow; it would take so much time to do manually.
To find out more about Tecan’s hit screening and lead optimization solutions, visit lifesciences.tecan.com/screening_and_lead_optimization
To learn more about AI and non-target drug discovery, go towww.transilico.com
Keywords:









