Keywords:
Live cell imaging is one of the most important techniques in life sciences today. But behind every great imaging assay, pity the poor scientist grappling with the demands of biological variability and complex kinetic assays. Live cell experiments are often synonymous with unsociable working hours, tedious protocols and unrepeatable results. In this blog, we explore what it takes to tame automated cell imaging assays, and take back control of kinetic experiments to get reliable results more quickly, with fewer errors and less aggravation.
Real-time cytometry is empowering life science exploration
The popularity of imaging living cells continues to grow year on year. In the last five years alone, there have been nearly 8,000 scientific publications on live cell imaging – more than in the preceding 40 years combined. Why? Because the old adage ‘seeing is believing’ is actually true: images convincingly capture aspects of living cells’ behavior and function that are otherwise difficult or impossible to detect.
Today’s cell imaging platforms enable real-time, non-destructive capture of individual cellular events, in multi-well formats that support higher throughputs and robust, cell-by-cell statistical analyses. When image-based cytometry is combined with the capabilities of a multimode plate reader, even more insights are possible. With multiple detection modalities, including both top- and bottom-reading configurations, you can multiplex more types of assays and make use of novel fluorescent probes to quantify complex dynamic events.
Why live cell experiments get out of hand
Real-time live cell assays often involve sensitive cell types and complex protocols that can easily go awry, wasting valuable resources and possibly giving you misleading results. Here are six common pitfalls that can send your live cell experiments spinning out of control:
1) Starting with unhappy cells
Many live cell assays are doomed to fail before they even start, because the cells are not in the exponential growth phase, or have been subjected to stress prior to the experiment – for example, when transferring from the tissue culture incubator to the slightly different environment in the imager or plate reader.
2) Experimental complexity
Kinetic live cell assays can get very complex, very quickly. That’s because you need to factor in many different fluorescent probes, time points, dose concentrations, replicates, control wells, cell types, and so on. The more elaborate the experiment, the more chances there are for variability to creep in and mistakes to be made. To make matters worse, manual steps and complicated instrument control software can significantly increase the likelihood of human error. Unfortunately, some experimental complexity is often unavoidable – you really do need all those concentrations, replicates and time points! Fortunately, cell imaging platforms are advancing to help us accommodate the necessary complexity, rather than avoid it.
3) Failing to adapt to biological variability
Living cells are inherently variable, which means that, to obtain consistent assay results from day-to-day or lab-to-lab, you may need to adjust experimental protocols and timings on the fly. For example, rather than always starting your assay a fixed number of hours after seeding, you may get more consistent results if you wait a variable time period until the cells reach their optimal confluence – say 80 percent. But what do you do if the cells reach 80 percent confluence when you aren’t in the lab? Quite often, we compromise and start the assay at the wrong confluence, rather than go back to the drawing board and rework the timings.When you compromise protocol timings to fit your schedule, the assay may still work, but reviewers may question whether your results are biologically relevant. In the example discussed later, we see how variable factors like percentage cell viability and fluorescent signal may dictate the most appropriate times to add various stimuli or probes, or when to start and stop imaging to ensure you capture the full response.
4) Disturbances and disruptions
Mechanical or environmental disturbances can trigger cellular stress responses that will alter cell behaviors and responses to stimuli, and may even activate cell death pathways. Common culprits include lid lifting, harsh addition of reagents or compounds, wash steps and transfer of plates from one device to another.
5) Scale-up and miniaturization
Kinetic assays may work fine at the prototype stage, only to fall prey to mistakes when scale-up suddenly amplifies the number of replicates, pipetting steps, samples and compounds. In addition, scale-up often entails miniaturization, which means dealing with very small liquid volumes. In such cases, evaporation – and, by extension, assay duration – becomes a significant concern. At the same time, liquid handling accuracy and precision become even more critical, and the chances of mistakes and cross-contamination are further magnified.
6) Image management challenges
Depending on the assay complexity, a live cell imaging experiment may generate thousands – or even hundreds of thousands – of images per run. Data storage and archiving can be expensive, and servers quickly become filled to capacity. If storage space runs out in the middle of a critical run, you risk losing the cells and the entire data set – a big toll in terms of the cost, time and labor you put into your experiment. For all these reasons, limiting the number of images you need to acquire can be a smart move.
When a simple assay isn’t so simple
Live cell imaging assays require a high degree of responsive control; the person running the experiment typically needs to monitor conditions frequently, and take action based on cell appearance or some other biological cue, such as intensity of a fluorescent indicator. To illustrate the challenges of live cell imaging assays, and the level of experimental control needed to avoid the pitfalls discussed above, let’s take a look at a relatively simple ‘live/dead’ cell viability assay. The goal of this assay is to monitor changes in percentage cell viability in response to varying doses of test compounds, generating a dose response curve and half maximal viability (IC50) for each. To assess viability, we are using a conventional combination of two fluorescent dyes: calcein AM to identify live (metabolically active) cells, and propidium iodide (PI) to detect dead cells (loss of plasma membrane integrity).
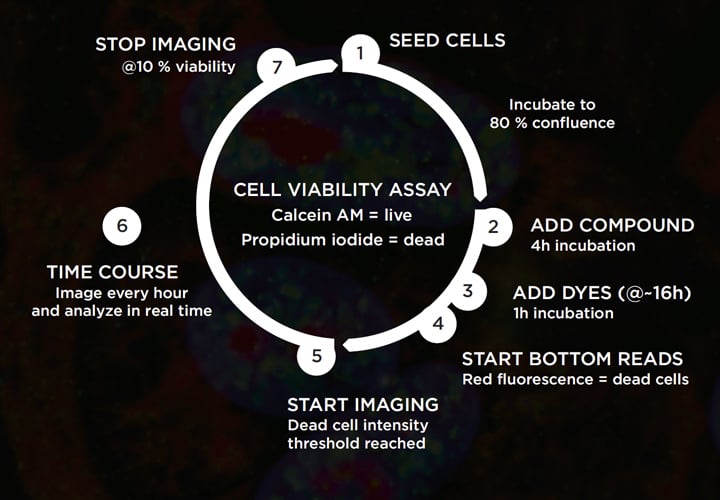
Seems pretty straightforward, right? But take another look at the assay schematic. There are several key points in the protocol when biological conditions trigger a particular action. If you are running the assay without full automation, you will need to assess the cells periodically on a manual microscope to monitor confluence and decide when to add compounds. The timings for compound addition, dye addition and PI fluorescence recording are all dictated by the confluence of the cells. Likewise, the ideal imaging start and stop times depend on when the cells start to be affected by each compound, and when the percentage cell viability bottoms out. Orchestrating all these actions manually or across multiple detection platforms without making any mistakes is not easy. Maybe this ‘simple’ live/dead assay is not so simple after all!
How to take back control of your live cell experiments
Let’s take a look at some improvements we can make to gain more control and eliminate errors.
1) Get on-board environmental control
At the start of the experiment, cells are seeded into a 384-well plate, and left to incubate until they have recovered and reached 80 percent confluence. Crucially, you need to avoid shocking the cells when transferring them from the tissue culture incubator to the liquid handling workstation or detection system. This means providing the cells with a stable, humidified on-board environment (typically 37 oC and 5 % CO2), where they can equilibrate for a sufficient amount of time before compounds are added and measurements are taken.
2) Automatically trigger actions using thresholds
Depending on the health of the cells at the time of seeding, the length of time needed to reach 80 percent confluence can vary unpredictably. To avoid the hassle of having to adapt your protocol and working hours to suit the cells, the smart solution is to automate confluence determination, and set up a confluence threshold that will automatically trigger subsequent steps. An automated multimode reader and cell imaging platform with this sort of conditional real-time experimental control can minimize the amount of manual intervention needed, while eliminating subjective assessments – triggering additions and reads at the right times, based on accurate quantitative measurements.
3) Reduce the number of images acquired
As shown in the schematic, the assay is configured to run for approximately 48 hours from start to finish. If we run the assay in just one 384-well plate, collecting a single whole-well image for both the red and green channels every hour from the point that the dyes have been equilibrated (ie. hours 17 through 48), we’ll need to collect a jaw-dropping 24,576 images. Can we do better? Of course. It turns out that we are collecting many images that are not needed, namely those at the very top and bottom of the dose response curves. The key to reducing the image number is to use threshold-based conditional programming to collect only the images that are actually needed to generate a reliable curve. Here’s how it works... After the cells have been equilibrated with the dye mix, we start continuously recording PI intensity using bottom reading (no images required!). We can then automatically detect an increase in fluorescence when dead cells first start appearing, and use an intensity threshold to trigger the two-color imaging to start from that point onward. Similarly, if we can calculate percent viability by analyzing the image data in real time, we can set a second threshold that will trigger the imaging to stop when viability bottoms out, in this case at 10 percent. With both thresholds applied, imaging is performed only from hours 20 to 44. The result? Automated real-time experimental control reduces the number of acquired images by 5,376 – over 20 percent! That’s a major saving in terms of both experiment time and storage space.
4) Create a walkaway process
A final consideration is the amount of hands-on time needed to run the assay from start to finish. As mentioned earlier, the more manual steps you can eliminate the better. Automating as much of the assay protocol as possible will significantly improve accuracy, decrease variability, reduce the chance of errors and free up valuable staff for more important, less tedious tasks.
The answer? A multimode imager with real-time experimental control
The best way to achieve hands-free, error-proof kinetic cell assays is with a multimode reader that includes on-board environmental control and lets you perform image-based confluence assessment, intensity measurements, fluorescence imaging and real-time image analysis – all on the same system. And crucially, you need a system that allows you to set up threshold-based conditional responses easily and with no need for programming skills.
Keywords:









