By Oliver Schmidt
How the human body deals with infection depends on an individual’s immune response. When looking at the body’s response to SARS-CoV-2, the state of the immune system has a crucial impact on the clinical outcome. For example, HMGB1 (High Mobility Group Box 1) protein is a key mediator of the immune system, and as such it has been shown to be critical in the replication of SARS-CoV-2. This article outlines the potential roles of HMGB1 in the race to find solutions to the coronavirus pandemic.
The immune system fights viral infections in the body. Visual representation of leukocytes attacking a COVID-19 virus particle.
HMGB1 and the immune response
Human HMGB1 is essential in the cell’s response to stress, and plays a major role in many disease states, including but not limited to infectious diseases, ischemia, immune disorders, neurodegenerative diseases, and cancer.1
The severity of the ongoing coronavirus pandemic has led to an exponential rise in research into COVID-19, and the most recent results and reviews suggest that HMGB1 plays a key role in regulating the inflammatory response in COVID-19 pathology.2-5
Two major areas of interest have come to light, which we will examine here:
- the role of HMGB1 in the replication of SARS-CoV-24
- a potential therapeutic role for HMGB1 in COVID-19 treatment 5
Since HMGB1 is pivotal in regulating the body’s immune response to infection, one can also imagine that its role in COVID-19 pathology is particularly important when infected patients already have underlying health conditions (co-morbidities) such as hypertension, diabetes, coronary heart disease, and cancer, where HMBG1 is known to exert its influence. In cancer, for example, HMGB1 has a role both as a tumor suppressor and as an inducer in oncogenesis. Now, it just so happens that as many as 94% COVID-19 deaths are amongst patients with co-morbidities, so what exactly might be happening here, and could HMGB1 have a role to play in the fight against the current pandemic?1, 6,7
Some answers can be found in our previous article, which outlines the fascinating science behind HMGB1 and its crucial place as a mediator in health and disease. However, for more specific answers that could help us find out what role human HMGB1 plays in SARS-CoV-2 infection and eventual clinical outcomes, we turn first of all to recent research that highlights the importance of HMGB1 in SARS-CoV-2 replication.
HMGB1 and SARS-CoV-2 viral replication
As HMGB1 is so critical to the immune response, it follows that if we can find out more about its potential role in COVID-19 infection, we may reveal novel therapeutic targets and contribute towards our understanding of COVID-19 pathogenesis. To this end, Wei et al. recently performed a genome-wide CRISPR screen to find potential host genes essential for SARS-CoV-2 infection.4
Viral entry, the first stage of the SARS-CoV-2 lifecycle, is mediated by the viral spike protein. The receptor binding domain of the spike protein binds to the cell surface receptor: Angiotensin-Converting Enzyme 2 (ACE2). The coronavirus spike protein then undergoes two proteolytic processing steps prior to entry. Upon viral membrane fusion, the viral RNA is released into the cytoplasm, where it is translated, and establishes viral replication and transcription complexes before assembling and budding.
Genome-wide CRISPR screening is an effective way to identify the host genes necessary for the individual steps involved in this infection process.
Hence, such screening can potentially be used to inform mechanisms of COVID-19 pathogenesis, reveal variation in host susceptibility, and identify novel host-directed therapies, which may have efficacy against current and future pandemic coronaviruses.
Using CRISPR screening, Wei et al. successfully identified known SARS-CoV-2 host factors, including the receptor ACE2 and protease Cathepsin L. In addition, they discovered novel pro-viral genes and pathways, including the SWI/SNF chromatin remodeling complex and key components of the TGF-β signaling pathway, and found that small molecule inhibitors of these pathways prevented SARS-CoV-2-induced cell death.
A major conclusion of their research was that HMGB1 is critical for SARS-CoV-2 replication. In contrast, loss of the histone H3.3 chaperone complex sensitized cells to virus-induced death. Therefore, this powerful study has already revealed potential therapeutic targets for SARS-CoV-2, as well as highlighted host genes that may regulate COVID-19 pathogenesis.
Another piece in the HMGB1 jigsaw puzzle
COVID-19 exhibits a heterogeneous clinical appearance. Whilst most patients are asymptomatic or exhibit mild to moderate symptoms, approximately 15% progress to severe pneumonia and about 5% are eventually admitted to the intensive care unit (ICU) due to acute respiratory distress syndrome (ARDS), septic shock and / or multiple organ failure. ICU patients respond poorly to currently available treatments and exhibit a high mortality rate. Inadequate identification of the determinants of fatal outcomes is one of the major obstacles to the improvement of the outcomes in severe COVID-19 patients.
It has been hypothesized that the alarmins S100A8/A9 and HMGB1 are significantly involved in COVID-19 outcomes, but until recently no studies had demonstrated this.
Chen et al. (2020) thus set out to define the role of S100A8/A9 and HMGB1 in progression to fatal outcomes and develop clinically relevant risk strata for COVID-19 patients.2
Their findings were significant. COVID-19 patients treated in general wards had elevated levels of S100A8/A9 (P = 0.033) but not HMGB1 (P > 0.9999) as compared to healthy controls, suggesting that S100A8/A9 is a more sensitive alarmin than HMGB1 in response to SARS-CoV-2 infection. However, both S100A8/A9 and HMGB1 had much higher extracellular levels in ICU-admission patients compared to non-ICU patients. Findings were similar in patients with fatal outcomes compared to patients that survive, indicating that significant elevation of S100A8/A9 and HMGB1 was associated with high mortality.
When looking at the serum levels of S100A8/A9 or HMGB1, 12 cytokines simultaneously correlated with both the S100A8/A9 and HMGB1. Interestingly, 3 myeloid chemokines, IL-8, MCP-3 and MCP-1, were among the most significant cytokines that simultaneously correlated with both the S100A8/A9 and HMGB1 and showed the lowest P value (P < 0.0001), indicating that the overproduced S100A8/A9 and HMGB1 in serum were associated with distinct signatures for cytokine storm in patients with COVID-19.
Can HMBG1-based therapeutics help cure COVID-19?
Exposure to hyperoxia, via mechanical ventilation with potentially toxic amounts of oxygen, such as performed in ICU with ARDS patients, results in accumulation of high levels of airway HMGB1 that precede inflammatory acute lung injury (ALI).8 Airway HMGB1 is critical in mediating hyperoxia-induced inflammatory ALI via recruiting leukocytes, including neutrophils. So essentially, the higher the level of extracellular HMGB1, the greater the likelihood of lung injury.
Entezari et al. (2020) were able to demonstrate that the inhibition of extracellular HMGB1 attenuates hyperoxia-induced inflammatory acute lung injury.8 They found that ethyl pyruvate (EP), or neutralizing anti-HMGB1 antibodies, administrated even after the onset of hyperoxic exposure, can mitigate hyperoxia-induced inflammatory acute lung injury by inhibiting HMGB1 release into the extracellular milieu, thus paving the way for the development of HMGB1-based therapeutics.
HMGB1/RAGE/TLR4 in severe pulmonary inflammation
Many chronic inflammatory diseases are characterized by increased circulating HMGB1 levels.9 This could increase the risk of severe outcome in COVID-19 patients with inflammatory comorbidities. Excessive extracellular HMGB1 quantities cause tissue damage and organ dysfunction. Treatment with HMGB1-specific antagonists ameliorates inflammation and improves survival in many preclinical models of acute or chronic inflammatory diseases.10-13 However, therapy with HMGB1 specific antagonists has not yet been studied in clinical trials.
Influenza studies provide interesting insights into ongoing COVID-19 research. Influenza viruses are RNA viruses, like SARS-CoV-2. Both replicate in respiratory epithelial cells and cause necrotic tissue damage. Influenza-infected patients express elevated circulating HMGB1 concentrations associated with the development of severe pneumonia.14 Gene-deficient TLR4 as well as gene-deficient RAGE mice (RAGE: receptor for advanced glycation end-products) are partially protected from influenza-induced lethality.15 Whilst similar studies have yet to be carried out with SARS-CoV-2, successful pre-clinical treatment results using specific HMGB1-, TLR4- or RAGE-antagonists further support the central role of the HMGB1/RAGE/TLR4-axis in the pathogenesis of influenza infections, and combined anti-HMGB1 mAb and anti-viral treatment offer almost complete protection against lung injury.16
The role of HMGB1 in inflammation / immune response
Our final study looks at the possibility that SARS-CoV-2 might directly activate the NLRP3 inflammasome, resulting in the endogenous adjuvant activity needed to mount a proper adaptive immune response against the virus.3 It postulates that the heterogeneous response of COVID-19 patients could be attributed to individual differences in the ability to properly down-regulate NLRP3 inflammasome activation, which in turn correlates to the fitness of the immune system of the individual challenged by the virus.
In effect, patients with a reduced immune fitness can demonstrate a dysregulated NLRP3 inflammasome activity, resulting in severe COVID-19 with tissue damage and a cytokine storm. The authors sketched the outlines of five possible scenarios for COVID-19 in medical practice and described potential treatment options targeting dysregulated endogenous adjuvant activity in severe COVID-19 patients, giving them a potential path back to health (Figure 1).
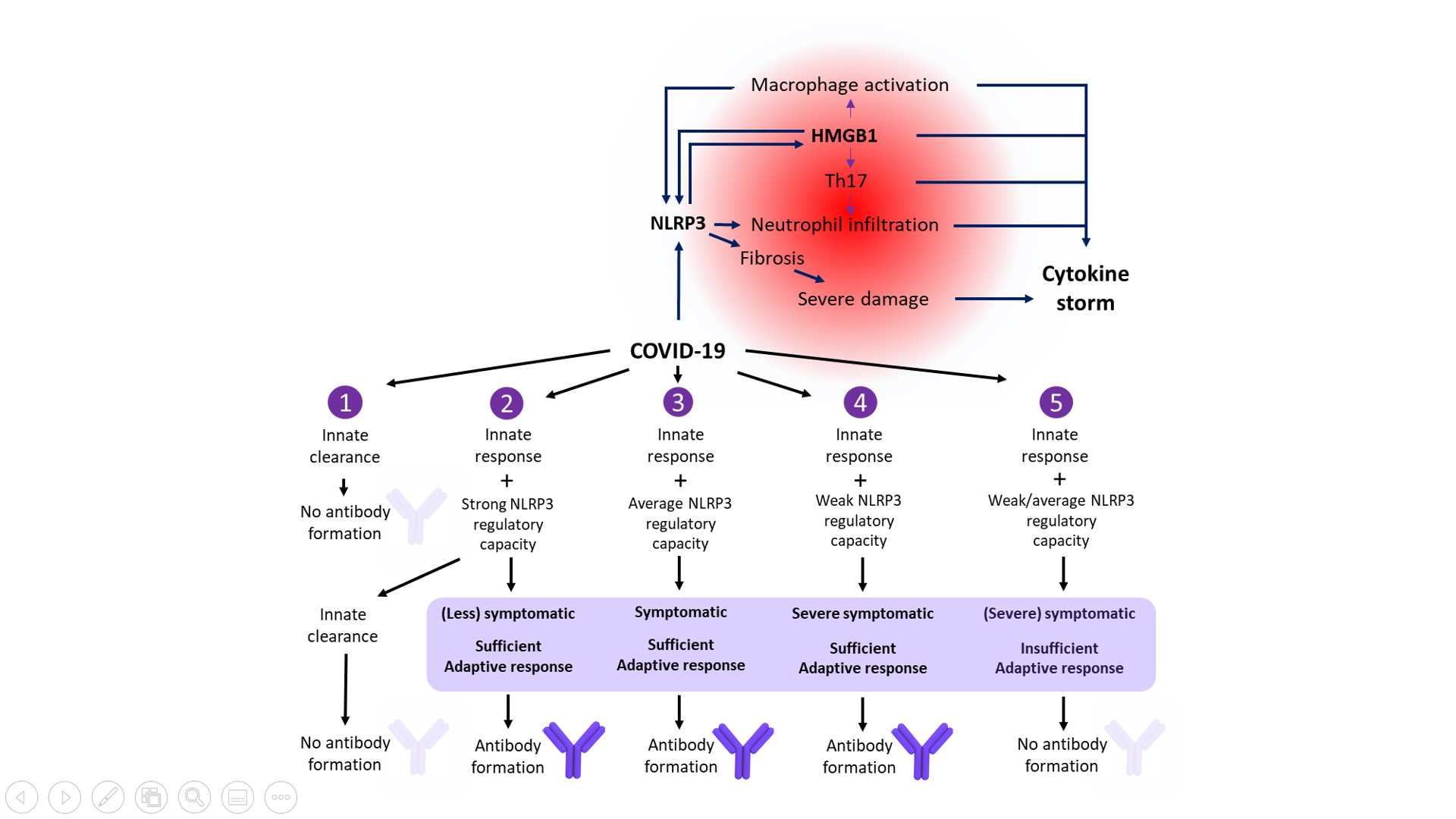
Figure 1. Central role of NLRP3 inflammasome activation and HMGB1 in the severe symptomatic phase of COVID-19. The clinical course of COVID-19 infection depends on an individual’s immune fitness. Five possible scenarios describing potential treatment options and potential paths back to health are shown. (Adapted from van den Berg and Te Velde, 2020).
If HMGB1 is so important, how do I measure it?
There is now significant evidence that HMGB1 is essential for SARS-COV-2 replication, as well as potentially being a therapeutic target in severe cases of COVID-19.17
It follows that labs must be able to measure HMGB1 accurately in serum and other samples, both to establish its level in different stages of SARS-CoV-2 infection, and to be able to use it as a key marker that can be monitored during treatment.
In the next article in this series we will examine methods to effectively measure HMGB1, such as the use of ELISA-based kits. Check out our poster to learn more about the complex and crucial role of HMGB1 in cell function.
References
1. Kang, R., Chen, R. et al. (2014). HMGB1 in health and disease. Molecular aspects of medicine, 40, 1–116. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/25010388/ DOI: https://doi.org/10.1016/j.mam.2014.05.001
2. Chen, L., Long, X. et al. (2020). Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cellular & Molecular Immunology, 17, 992-994. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/32620787/ DOI: https://doi.org/10.1038/s41423-020-0492-x
3. van den Berg, D.F. and Te Velde, A.A. (2020). Severe COVID-19: NLRP3 Inflammasome Dysregulated. Frontiers in immunology, 11, 1580. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/32670297/ DOI: https://doi.org/10.3389/fimmu.2020.01580
4. Wei J., Alfajaro M.M. et al. (2020) Genome-wide CRISPR screen reveals host genes that regulate SARS-CoV-2 infection. bioRxiv [Preprint]. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/32869025/ DOI: http://doi.org/10.1101/2020.06.16.155101
5. Andersson, U., Ottestad, W. and Tracey, K.J. (2020). Extracellular HMGB1: a therapeutic target in severe pulmonary inflammation including COVID-19?. Molecular medicine (Cambridge, Mass.), 26(1), 42. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/32380958/ DOI: https://doi.org/10.1186/s10020-020-00172-4
6. https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/evidence-table.html Accessed 15 October 2020
7.https://www.hopkinsguides.com/hopkins/view/Johns_Hopkins_ABX_Guide/540747/all/Coronavirus_COVID_19__SARS_CoV_2_Accessed 15 October 2020
8. Entezari, M., Mohammad, J. et al. (2014) Inhibition of extracellular HMGB1 attenuates hyperoxia-induced inflammatory acute lung injury. Redox Biol. 2, 314–322 PubMed ID: https://pubmed.ncbi.nlm.nih.gov/24563849/ DOI: https://doi.org/10.1016/j.redox.2014.01.013
9. McDonnell, M., Liang, Y. et al. (2011) Systemic toll-like receptor ligands modify B-cell responses in human inflammatory bowel disease, Inflammatory Bowel Diseases, Volume 17 (1), 298–307 PubMed ID: https://pubmed.ncbi.nlm.nih.gov/20806343/ DOI: https://doi.org/10.1002/ibd.21424
10. Andersson, U. and Tracey, K.J. (2011). HMGB1 is a therapeutic target for sterile inflammation and infection. Annual review of immunology, 29, 139–162. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/21219181/ DOI: https://doi.org/10.1146/annurev-immunol-030409-101323
11. Kang, R., Chen, R. et al. (2014). HMGB1 in health and disease. Molecular aspects of medicine, 40, 1–116. PubMed: https://pubmed.ncbi.nlm.nih.gov/25010388/ DOI: https://doi.org/10.1016/j.mam.2014.05.001
12. Andersson, U., Yang, H. and Harris, H. (2018). Extracellular HMGB1 as a therapeutic target in inflammatory diseases. Expert opinion on therapeutic targets, 22(3), 263–277. PubMed: https://pubmed.ncbi.nlm.nih.gov/29447008/ DOI: https://doi.org/10.1080/14728222.2018.1439924
13. Yang, H., Wang, H. and Andersson, U. (2020). Targeting Inflammation Driven by HMGB1. Frontiers in immunology, 11, 484. PubMed: https://pubmed.ncbi.nlm.nih.gov/32265930/ DOI: https://doi.org/10.3389/fimmu.2020.00484
14. Ito, Y., Bhawal, U.K. et al. (2012). Involvement of HMGB1 and RAGE in IL-1β-induced gingival inflammation. Archives of oral biology, 57(1), 73–80. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/21861984/ DOI: https://doi.org/10.1016/j.archoralbio.2011.08.001
15. van Zoelen, M.A., Yang, H. et al. (2009). Role of toll-like receptors 2 and 4, and the receptor for advanced glycation end products in high-mobility group box 1-induced inflammation in vivo. Shock (Augusta, Ga.), 31(3), 280–284. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/19218854/ DOI: https://doi.org/10.1097/SHK.0b013e318186262d
16. Hatayama, K., Nosaka, N. et al. (2019). Combined effect of anti-high-mobility group box-1 monoclonal antibody and peramivir against influenza A virus-induced pneumonia in mice. Journal of medical virology, 91(3), 361–369. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/30281823/ DOI: https://doi.org/10.1002/jmv.25330
17. Street M.E. (2020). HMGB1: A Possible Crucial Therapeutic Target for COVID-19?. Hormone research in paediatrics, 93(2), 73–75. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/32375153 DOI: https://doi.org/10.1159/000508291
About the author

Oliver Schmidt
Oliver Schmidt is product manager for research reagents, neurodegeneration, and bone and mineral products at TECAN-IBL in Hamburg with more than 10 years’ experience in bringing these products to the Life Science and academic community. He studied Environmental Engineering at the Technical University of Hamburg-Harburg and joined TECAN-IBL in 2005.











