By Beatrice Marg-Haufe
One of the steps in DNA sample preparation that is often overlooked when moving from manual to automated methods, is the quantification and normalization of nucleic acid samples that are destined for downstream analysis in different techniques and applications such as genotyping and NGS.
We saw in our third blog how to overcome the more obvious automation challenges associated with software and worktable design, and how to ensure that your automation protocols, or scripts, are optimized accordingly.
In this fourth and final blog of the series, we look in more detail at the practical benefits of incorporating nucleic acid quantification and normalization steps into your automated workflow.
.jpg?width=768&name=Quant_Norm_Normalization_workflow_Card_720x370%20(1).jpg)
Automated quantification and normalization of nucleic acid samples in genomics applications. The Freedom EVOware® Normalization Wizard is a software component that efficiently automates pipetting steps for the quantification and normalization of nucleic acid samples.
Challenges for automating nucleic acid quantification and normalization
DNA quantification and normalization workflows typically include the following steps:
- Set-up of quantification assay
- Determination of the DNA concentration
- Calculation of a dilution scheme to achieve the target concentration for each well
- Dilution of the DNA samples to the desired concentration
There are many questions and considerations when integrating these steps into your automated workflow, some of which are outlined below:
1. Are you ready to move from manual to automatic?
It is not uncommon for labs to automate nucleic acid extraction and purification, but then revert to fastidious and tedious manual methods for quantification and normalization of their precious samples prior to downstream analysis.
Proposals to automate these remaining manual steps often meet with resistance because the techniques have been established over many years, and disturbing the status quo seems overly risky, inconvenient or expensive. However, any downside to moving to a fully automated workflow is usually greatly outweighed in the long run due to improved cost-efficiency and decreased error rates.
2. Have you got enough DNA?
When working with irreplaceable samples and/or small sample volumes, the amount of DNA available for downstream analysis is limited, so any workflow improvement that increases efficiency and reliability can be a real game changer. Automation provides maximum sample economy, in many cases enabling you to move to smaller pipetting volumes than you could reliably and accurately handle manually. However, the flexibility of your automation solution is key when working with low or variable yields because protocol modifications are often needed to get the most out of the available sample. For example, if your DNA yield is exceptionally low, you may decide to move quantification from a 96-well to a 384-well format, decrease the number of replicates, or have your system ask for a user-specified command when the system comes across a sample out of predefined concentration ranges.
3. Are you quantifying your DNA accurately?
Whilst there is obviously a big difference between manual and automated quantification and normalization, the basic techniques remain the same. Two of the most common are determination of UV absorbance using spectrophotometry, and quantification of PicoGreen®, an intercalating fluorescent probe specific for double stranded DNA (dsDNA).
Quantification and normalization of dsDNA are prerequisites for a wide range of genomics applications, such as next-generation sequencing and quantitative PCR. The introduction of PicoGreen and other fluorescence-based probes has made selective quantification of even small amounts of dsDNA possible, overcoming the limitations of classical absorbance techniques.1
4. Can you pipette and calculate consistently?
When performing quantification and normalization for downstream techniques like NGS, PicoGreen quantification in a 384-well plate format is typically used. An experienced user will be able to successfully and reliably pipette the microvolumes which are required for this process. However, reliability is very user-specific, and not everyone will always get the same results manually.
Moreover, with manual quantification, researchers need to calculate from the raw data how to normalize their sample concentrations. Following quantification, the user will usually have a complex Excel® sheet, from which they will calculate how much sample and how much diluent to add to add to each well. With every single sample potentially requiring a different dilution factor, the chances of making a mistake can be high. Automating both the calculations and the dilution process significantly reduces the likelihood of errors.
PicoGreen quantification and normalization with DreamPrep™ NAP
To address the above challenges a fully automated “walk away” sample prep automation solution has been developed, DreamPrep NAP. This solution includes nucleic acid extraction as well as optional quantification and normalization and is compatible with the Quant-iT PicoGreen dsDNA Assay Kit, making it a seamless process with excellent reproducibility. DreamPrep NAP combines the Fluent® Automation Workstation, an Infinite® 200 Pro multimode plate reader in M Nano+ configuration, and Fluent GX Assurance Software.
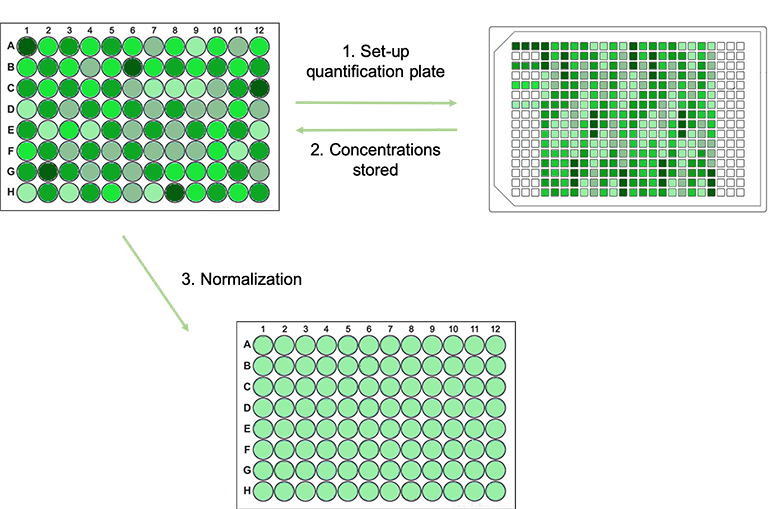
With a fully integrated quantification and normalization script on board, all the user has to do is enter the required post-normalization DNA concentration. All subsequent plate preparation steps, dilutions, measurements and calculations are then carried out automatically (Figure 1).
Figure 1. Schematic workflow of the Fluent quantification and normalization protocol. (1) Samples prediluted (not shown) and pipetted into the quantification plate. (2) Concentration of each sample measured and stored. (3) Samples normalized in a new microplate.
The Fluent Laboratory Automation Solution and Fluent GX Assurance Software software simplify your workflow and eliminate the need for manual calculations, while FluentControl™ Smart Commands give you the flexibility you need for DNA normalization:
Store Well Concentration – records the concentration of each sample, taking into account potential dilution steps.
Normalization – uses saved data to automatically generate a script to achieve the desired concentration and volume for each sample, either in situ or in a new plate.
Fluent’s user-friendly TouchTools™ touchscreen interface guides the operator through the various selection options (sample number, number of replicates, desired final concentrations and target volumes), and provides detailed guidance for instrument set-up, ensuring adherence to SOPs and increasing process security for more consistent results.
Minimal cross-contamination, high reproducibility
To assess performance, the quantification and normalization workflow was tested on experimental samples. The samples and normalization assays were handled in 96-well format, and the quantification carried out in 384-well format. The worktable was configured, and parameters were selected as prompted by the FluentControl software. Full experimental details can be found in our application note.2
The system is able to perform the whole quantitation and normalization workflow in just 80 minutes – 55 minutes for quantification plate set-up, 10 minutes for reading and 15 minutes for normalization – including sample dilution based on the typical yields for the intended extraction workflow.
To assess the degree of cross-contamination, samples at high and low concentration were distributed in a checkerboard configuration across the 384-well plate. The results ruled out any significant cross-contamination by demonstrating that there was no significant variation in any part of the plate (Figure 2).
Following quantification of sample concentration, the samples from the checkerboard plate were automatically normalized to achieve a target concentration of 2.5 ng per µl. After the normalization step, the concentration in each well was measured again to assess repeatability. The average concentration of the samples was found to be on target at 2.6 ng/µl, with a tight CV of 6.9% (Figure 3).
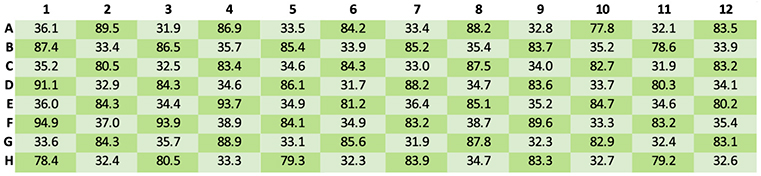
Figure 2. Quantification results for the assessment of cross-contamination using the automated method (Darker shading = average concentration 84.7 ng/µl; Lighter shading = average concentration 34.1 ng/µl).
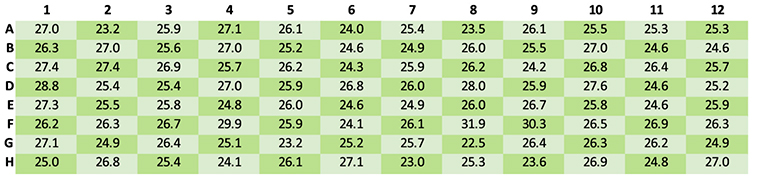
Figure 3. Quantitation results for samples normalized to 25 ng/µl using the automated method. The observed average concentration was 25.9 ng/µl, with a CV of 5.5%.
DreamPrep NAP: automation from start to finish
The consistency in these experimental results shows that DreamPrep NAP does indeed deliver a viable automated workflow solution, allowing seamless transition between the liquid handler and the reader. Only a single provider is needed for the full automation solution, and all associated calculations are performed on-board, without any need to integrate third-party solutions.
With the reproducibility that automation can deliver, DreamPrep NAP helps ensure that you are not studying accidentally introduced artefacts, and that you are focusing on real inter-sample differences. Contact us now to find out more about how DreamPrep NAP can help you move your research goalposts one automated step further towards publication and future implementation in clinical applications.
Contact Tecan to find out more about automated workflows for DreamPrep NAP
References
- Singer V.L., Jones L.J., Haugland R.P. (1997) Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal. Biochem. 249:228–238.
- Tecan Application Note: Automation of the Quant-iT™ PicoGreen® quantification and normalization of double stranded DNA with minimal set-up time. dsDNA Assay Kit on the Fluent®, Application Note, Laboratory Automation Solution
About the author

Dr Beatrice Marg-Haufe
Dr. Beatrice Marg-Haufe is a product manager at Tecan Switzerland with over 10 years of experience in assay development and product management. She studied biochemistry at the University of Bielefeld, Germany, and at Harvard Medical School, USA. She focused on cancer research during her PhD in Biochemistry at the MPI, Munich, Germany. She joined Tecan in 2009 focusing on applications for the agriculture and genomics market.











