By Oliver Schmidt
In the first article in this series, we looked at how HMGB1 has taken an increasingly important position as a key mediator in the immune response, playing a major role in many diseases, from cancer to coronavirus. There is now significant evidence that HMGB1 is essential for SARS-COV-2 replication, as well as potentially being a therapeutic target in severe cases of COVID-19.1 In this article we examine how we can effectively measure HMGB1 accurately in serum and other samples and begin the journey from research to clinic.
 HMGB1 is implicated in the immune response to SARS-CoV-2
HMGB1 is implicated in the immune response to SARS-CoV-2
HMGB1: redox state matters
In the past, the usefulness of HMGB1 as a prognostic/diagnostic biomarker was hampered by the need to understand the isoforms that control different functions, and the absence of robust methods for measuring it. The different isoforms result from post-translational modifications, with the redox states of three cysteines at positions 23, 45 and 106 critical for activity. Depending on the redox states of these residues, HMGB1 can induce cytokine production via toll-like receptor 4 (TLR4) or promote chemotaxis by binding the chemokine CXCL12 for stimulation via CXCR4. Fully oxidized HMGB1 is inactive. These different redox states are summarized in Figure 1.
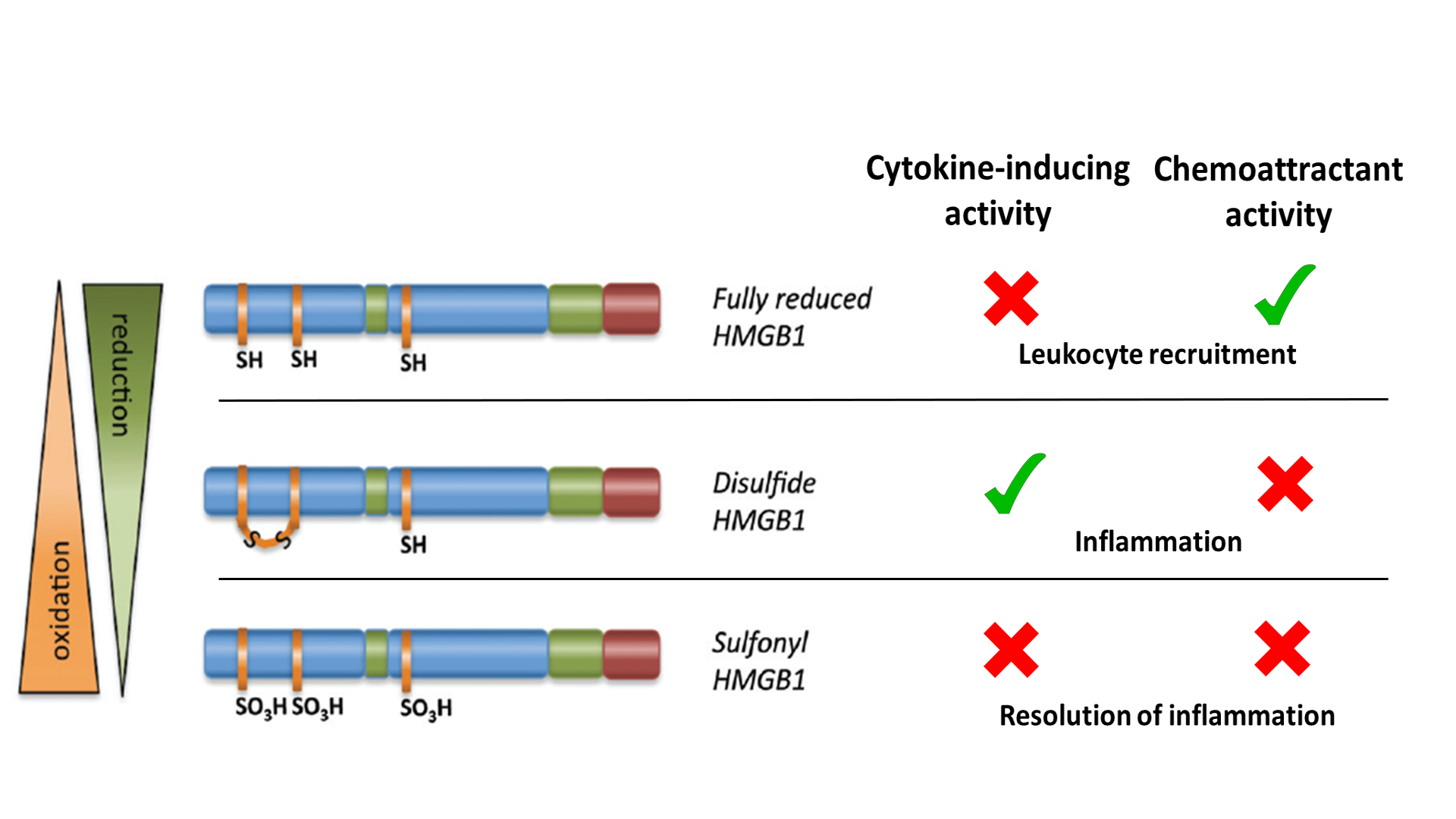
Figure 1. Summary of HMGB1 redox state versus activities (adapted from Venereau et al., 2012 and Tang et al., 2012).2,3
To act as a DAMP/danger signal and inflammatory mediator, HMGB1 is transported extracellularly by two principal means: active secretion from living inflammatory cells (for example, macrophages) or passive release from necrotic cells. The activities of extracellular HMGB1 are redox dependent. All-thiol-HMGB1 promotes chemokine production and leukocyte recruitment. Disulfide-HMGB1, originating from infiltrating leukocytes, promotes release of proinflammatory cytokines and thus participates in the inflammatory response. Reactive oxygen species produced by leukocytes induces the terminal oxidation of HMGB1, which is inactivated during resolution of inflammation.
Taken together, this means that HMGB1 can play multiple roles in the pathogenesis of inflammatory and autoimmune disease, and mediate processes that range from inflammation to repair.4
Measuring HMGB1: the vision
HMGB1 is routinely included as a necessary biomarker in consensus guidelines for the detection of immunogenic cell death.5 However, the journey towards its meaningful measurement has been a rocky one, complicated by such distractions as the need to resolve the different isoforms, and to overcome interference caused by auto-antibodies and other proteins that naturally interact with HMGB1 to modulate its function.
Western blotting has been traditionally included in the detailed analysis of protein structure and function, but whilst it can be used to analyze the different isoforms of HMGB1, the fact that it is comparatively laborious and only semi-quantitative when screening more than a few samples meant that researchers naturally turned to ELISA as an alternative.
ELISA to the rescue?
Developing an ELISA to measure HMGB1 would offer clear advantages over western blotting, including ease of use and the high throughput envisaged, as the critical role of HMGB1 began to unfold across the disease spectrum, most recently in the case of the coronavirus pandemic, where it has been identified as a key player in SARS-CoV-2 replication.
The challenge was huge: to develop the antibody reagents and sensitive assays for a protein that is so central to the mammalian immune response, with a range of modifications that are critical to its function. Creating effective antibodies against HMGB1 sounds like a fight against nature.
As Marco Bianchi, a leading scientist in the HMGB field and founder of HMGBiotech (www.hmgbiotech.eu) puts it:
“HMGB1 is one of the pillars of the immune system, so whenever you try use the immune system to create reagents against HMGB1 you are asking the immune system to make antibodies against one of the main cogs in the immune machinery.”
Assay developers usually aim to use highly specific monoclonal antibodies in ELISA, but it has been a fifteen-year struggle just to develop the three monoclonal antibodies that exist so far, and they all have a low affinity for HMGB1. Getting ELISAs based on monoclonal antibodies onto the market is a clear goal for suppliers.
Meanwhile, the resolution of the leading ELISA for total HMGB1 is still based on polyclonal capture antibodies raised in chickens, although the detection antibody, which identifies both HMGB1 and HMGB2, is monoclonal.6 Birds have a slightly different HMGB1 with a few amino acid changes and mammalian HMGB1 is better recognized by the avian immune system. Whilst the development of a monoclonal detection antibody represents a significant step forward, HMGB1 ELISA technology would still benefit from the development of a monoclonal antibody that is highly specific for HMGB1. Any decrease in the overall length of the assay would also be advantageous, since it currently relies on an overnight incubation.
The challenge of interference
Even high-performance ELISAs for HMGB1 are faced with challenges, including interference. In their pioneering work on ELISA interference, Vilma Urbonaviciute, at IZKF Research Group 2, Erlangen, Germany, and her co-workers found that ELISA detected lower levels of HMGB1 than immunoblotting.7 They suspected that the cause might be the IgG and IgM antibodies that bound to HMGB1 in most sera and plasma samples they investigated, including those from healthy individuals.
HMGB1 has such a ubiquitous role in the cell that it is perhaps not surprising that measurement in serum and plasma can be severely affected by natural interactions with a large range of molecules. For example, HMGB1-binding antibodies may well play an important role in biological functions, such as limiting overwhelming cytokine release as a result of tissue damage or sepsis, such as that seen in ICU patients with COVID-19.
Added to that, the activity of HMGB1 largely relies on its ability to form biologically active complexes with cytokines such as IL-1β, with lipopolysaccharide, or with CpG oligonucleotides. Interference can be particularly problematic in diseases that have a high prevalence of autoantibodies against HMGB1 in patients with autoimmune diseases, including systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA).8,9
To solve this interference problem in ELISA, Stéphanie Barnay-Verdier and her colleagues prepared in vitrocomplexes containing HMGB1 protein. Recombinant human HMGB1 (rhHMGB1) was incubated in the presence of molecules that are known to form complexes with HMGB1 such as LPS, IL-1β, or a rabbit antiserum directed against HMGB1. They then tested the capacity of perchloric acid (PCA) to dissociate these complexes by quantifying rhHMGB1 by ELISA immediately or following PCA treatment.10
They were able to demonstrate for the first time that incubation of rhHMGB1 with, IL-1β, LPS or specific antibodies significantly reduces the amount of protein detected by conventional ELISA (p<0.05). Treating the samples with PCA prior ELISA efficiently reversed this inhibition. PCA-modified ELISA was able to detect significantly higher amounts of HMGB1 in samples of plasma from 40 patients with septic shock compared to conventional ELISA (p=0.0006).10
The vision of detecting isoforms with ease
Despite all the advances made in ELISA, which remains the dominant method for total HMGB1 measurement, antibody development has yet to deliver the specificity required to resolve and measure isoforms resulting from post-translation modifications such as acetylation and oxidation that determine the location and function of HMGB1. Isoforms of HMGB1 can be identified by LC-MS but it offers extremely low throughput and is highly complex, so the search goes on for a way to detect isoforms, ideally using an ELISA-based method.11
As we unravel the intricacies of the role of HMGB1 as a major mediator in the immune response, including that for COVID-19, the translation of that research to the clinic would ideally be able to rely on a sensitive, easy-to-use and high throughput assay capable of resolving isoforms. The hope is that more specific antibodies can be generated, to enable the development of isoform-specific ELISAs. Meanwhile, robust, non-isoform specific ELISAs might at least represent a feasible compromise.
In the last of this three-part series, we will put ELISA technology to the test, and see how HMGB1 ELISAs are being used to dig deeper into the role of this fascinating group of proteins across a spectrum of diseases. We will thus see how it might be possible to establish the level of HMGB1 in different stages, samples and types of SARS-CoV-2 infection and potentially use it as a key marker that can be monitored during treatment. Meanwhile, check out Professor Ulf Andersson’s webinar where he explains the possible link between HMGB1 and COVID-19.
References
1. Street M.E. (2020). HMGB1: A Possible Crucial Therapeutic Target for COVID-19? Hormone research in paediatrics, 93(2), 73–75. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/32375153 DOI: https://doi.org/10.1159/000508291
2. Venereau, E., Casalgrandi, M. et al. (2012) Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release J. Exp. Med. 2012 Vol. 209 No. 9 1519-1528. PubMedID: https://pubmed.ncbi.nlm.nih.gov/22869893/ DOI: www.jem.org/cgi/doi/10.1084/jem.20120189
3. Tang, D., Billiar, T.R. & Lotze, M.T. A Janus Tale of Two Active High Mobility Group Box 1 (HMGB1) Redox States. Mol Med 18, 1360–1362 (2012). PubMed ID: https://pubmed.ncbi.nlm.nih.gov/23073660/ DOI: https://doi.org/10.2119/molmed.2012.00314
4. Magna, M., & Pisetsky, D. S. (2014). The role of HMGB1 in the pathogenesis of inflammatory and autoimmune diseases. Molecular medicine (Cambridge, Mass.), 20(1), 138–146. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/24531836/ DOI: https://doi.org/10.2119/molmed.2013.00164
5. Kepp, O., Senovilla, L., Vitale, I., Vacchelli, E., Adjemian, S., Agostinis, P., Apetoh, L., Aranda, F., Barnaba, V., Bloy, N., Bracci, L., Breckpot, K., Brough, D., Buqué, A., Castro, M. G., Cirone, M., Colombo, M. I., Cremer, I., Demaria, S., Dini, L., … Galluzzi, L. (2014). Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology, 3(9), e955691. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/25941621/ DOI : https://doi.org/10.4161/21624011.2014.955691
6. Yamada, S., Inoue, K., Yakabe, K., Imaizumi, H., & Maruyama, I. (2003). High mobility group protein 1 (HMGB1) quantified by ELISA with a monoclonal antibody that does not cross-react with HMGB2. Clinical chemistry, 49(9), 1535–1537. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/12928240/ DOI: https://doi.org/10.1373/49.9.1535
7. Urbonaviciute, V., Fürnrohr, B. G., Weber, C., Haslbeck, M., Wilhelm, S., Herrmann, M., & Voll, R. E. (2007). Factors masking HMGB1 in human serum and plasma. Journal of leukocyte biology, 81(1), 67–74. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/17060363/ DOI: https://doi.org/10.1189/jlb.0306196
8. Abdulahad, D. A., Westra, J., Limburg, P. C., Kallenberg, C. G., & Bijl, M. (2010). HMGB1 in systemic lupus Erythematosus: Its role in cutaneous lesions development. Autoimmunity reviews, 9(10), 661–665. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/20546955/ DOI: https://doi.org/10.1016/j.autrev.2010.05.015
9. Andersson, U., & Erlandsson-Harris, H. (2004). HMGB1 is a potent trigger of arthritis. Journal of internal medicine, 255(3), 344–350. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/14871458/ DOI: https://doi.org/10.1111/j.1365-2796.2003.01303.x
10. Barnay-Verdier, S., Gaillard, C., Messmer, M., Borde, C., Gibot, S., & Maréchal, V. (2011). PCA-ELISA: a sensitive method to quantify free and masked forms of HMGB1. Cytokine, 55(1), 4–7. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/21474328/ DOI: https://doi.org/10.1016/j.cyto.2011.03.011
11. Kwak, M. S., Kim, H. S., Lkhamsuren, K., Kim, Y. H., Han, M. G., Shin, J. M., Park, I. H., Rhee, W. J., Lee, S. K., Rhee, S. G., & Shin, J. S. (2019). Peroxiredoxin-mediated disulfide bond formation is required for nucleocytoplasmic translocation and secretion of HMGB1 in response to inflammatory stimuli. Redox biology, 24, 101203. PubMed ID: https://pubmed.ncbi.nlm.nih.gov/31026770/ DOI: https://doi.org/10.1016/j.redox.2019.101203
About the author

Oliver Schmidt
Oliver Schmidt is product manager for research reagents, neurodegeneration, and bone and mineral products at TECAN-IBL in Hamburg with more than 10 years’ experience in bringing these products to the Life Science and academic community. He studied Environmental Engineering at the Technical University of Hamburg-Harburg and joined TECAN-IBL in 2005.











